Fantastic Organelles and How to Build Them
Discover the power of synthetic organelles and how they can revolutionize biotechnology.
Fantastic Organelles and How to Build Them (Chapter 1)
Ever wondered what makes your cells work like mini factories, each with its own department handling different tasks? That’s where organelles come in. Think of them as specialized compartments inside a cell - like an office breakroom, but instead of coffee machines and snack drawers, they’re equipped to handle energy production (mitochondria and chloroplasts), DNA storage (nucleus), and waste management (lysosomes)[1]. Many organelles exist in almost all eukaryotes, from yeast and fungi to much larger plants and animals, including us humans.
Now, imagine trying to get bacteria to do those same tasks. The problem? Bacteria don't have breakrooms. They’re like open-floor offices with everything mashed together, making it tougher to specialize tasks. Although some bacteria have primitive organelles or other mechanisms for spatial organization [2], they lack the familiar structures of eukaryotic cells, like each open-floor office having its own, unique setup.
So, what if we could equip bacteria with their own versions of these toolkits, a synthetic organelle? In this blog, we’ll dive into why building synthetic organelles, especially in bacteria, is a game-changer.
What We Mean by “Organelles” (Let’s Get Specific)
First, let’s clear up a little confusion about the term organelle. Some definitions get blurry, lumping together everything from large molecular complexes like ribosomes, which every cell contains, to bacterial microcompartments. For our purposes, let’s focus on structures that:
Are micron-scale, meaning large enough to compartmentalize complex functions and often visible under an optical microscope.
Have the ability to selectively allow biomolecules (such as proteins or RNA) to come in and out. This is critical for efficient processing and regulation of biological reactions.
Include both membrane-bound organelles (like the nucleus or mitochondria) and membraneless organelles (like the nucleolus), which work their magic without a clear boundary but still manage to organize tasks.
We’re not talking about nanoparticle assemblies or molecular complexes here. Those are important, but for this discussion, we’re sticking to the bigger, function-compartmentalizing structures.
Why Bacteria?
Bacteria are the original minimalists. They’re small, efficient, and can perform a wide range of tasks - yogurt and vinegar, for example, are made from bacteria. The first recombinant biologic drug, insulin, was also produced using bacteria.
But here’s the catch - they’re usually missing the specialized compartments that eukaryotic cells rely on to stay organized, especially the bacterial strains that we often study in the lab and use in the factories. If you've worked in an open-floor office, you know the chaos: phones ringing, people typing, coffee-fueled conversations, and all the noise that interferes with your work. And if you need a stapler? Good luck finding it. It’s always on someone else’s desk instead of where it "belongs." In bacterial cells, there are 3-4 million proteins [3], and even more molecules floating around. Without compartments, it’s a mess!
For natural fermentation processes, bacteria are well-adapted to this complexity—thanks to billions of years of evolution. However, when it comes to engineering tasks, this lack of organization becomes a problem. Sure, you can get things done, but it’s inefficient - imagine trying to assemble IKEA furniture in a bustling lab where everyone is running their own experiments. Now picture trying to engineer a drug in a similarly chaotic bacterial environment.
What if we could give bacteria their own "breakrooms" for different tasks? These synthetic organelles would allow bacteria to compartmentalize functions, turning them into supercharged mini factories. With synthetic organelles, bacteria could do more, faster, and more efficiently.
A Brief History of the Attempts at Spatial Organization
Why has it taken so long to figure out how to give bacteria organelle compartments? As Richard Feynman famously said, "What I cannot create, I do not understand." Engineering biology often begins with learning from nature and then repurposing what we’ve found, or even creating entirely new design principles from scratch.
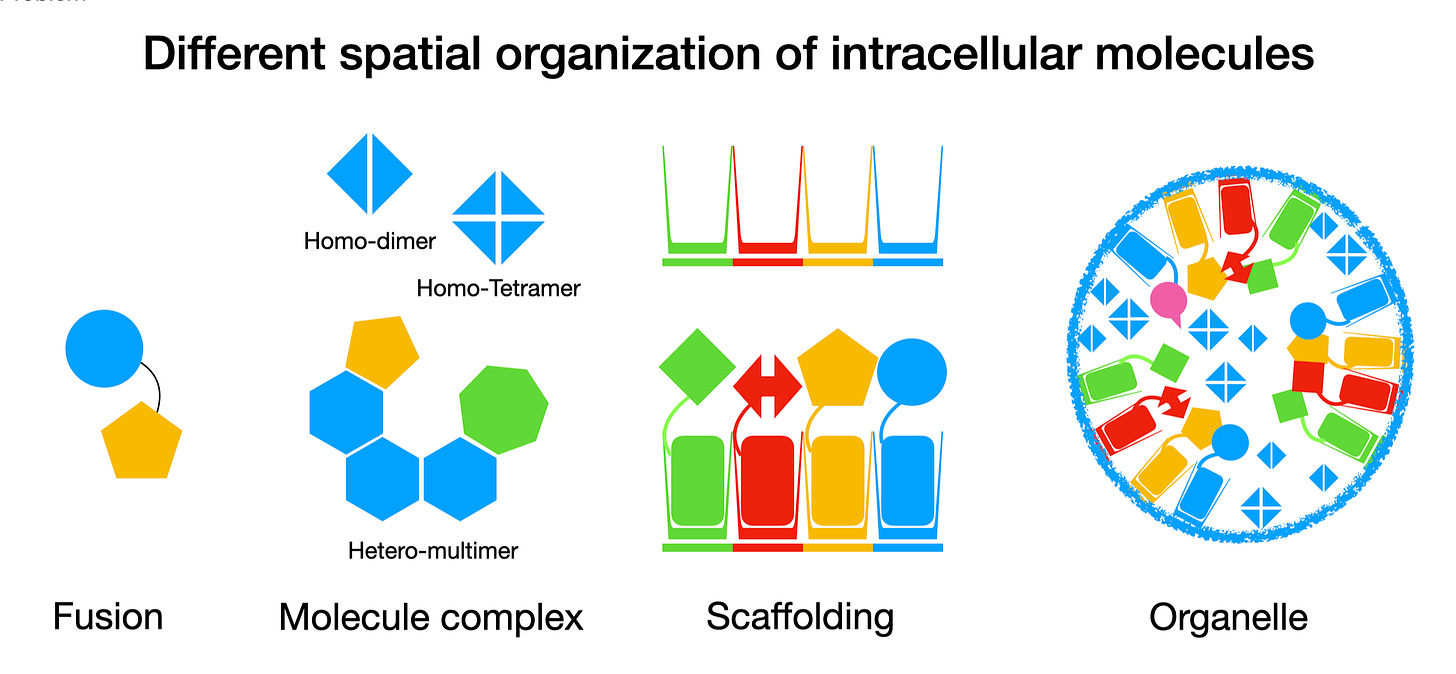
In nature, cells - especially eukaryotic cells - have mastered the art of spatial organization over billions of years. They use precise molecular assemblies and complex endomembrane systems. But nature hasn’t left us much of a blueprint, just genome sequences without clear annotations. As a result, early attempts at spatial engineering focused on mimicking simple, easy-to-understand spatial organization using fusion genes and self-assembly structures.
Fusion proteins, developed as early as the 1980s, were one of the earliest and simplest methods [4]. A direct approach, fusion proteins allowed scientists to co-localize two target proteins, binding them together in a predictable way. For instance, GFP (Green Fluorescent Protein) has been used as a fusion tag to track proteins under a microscope since its discovery [5]. Guided by mathematical models, fusion proteins have also been used to enable metabolic channeling, directing reactions to follow specific routes by forcing proteins to work in proximity [6].
By the 2010s, researchers began experimenting with molecular scaffolds to arrange molecules in pre-defined configurations, which led to some innovative designs. Examples include:
Protein scaffolds (pioneered by Keasling’s group) [7],
RNA scaffolds (pioneered by Silver and Lindner's groups) [8], and
DNA scaffolds (developed by Jerala’s group and Slovenia’s iGEM team, which won the grand prize in 2010) [9].
These methods achieved some success. By bringing specific enzymes into close proximity, molecular scaffolds improved the efficiency of metabolic pathways, regulated CRISPR/Cas systems for genome editing, and organized intracellular reactions. RNA scaffolds, in particular, provided a modular and programmable way to assemble proteins into a specific layout within the cell, capable of forming higher-order structures like filaments and granules at high concentration, mimicking organelle-like behavior [8].
But the limitations soon became apparent. Despite their flexibility and programmability, maintaining these self-assembling structures in the constantly shifting intracellular environment proved challenging. Scaffolded enzymes could swap between scaffolds, disrupting the stoichiometry and potentially decreasing overall efficiency [7]. Additionally, these scaffolds didn’t prevent interference between different reactions occurring in the same space due to their soluble nature. Even the best scaffolds weren’t able to create fully independent micro-environments like true organelles do.
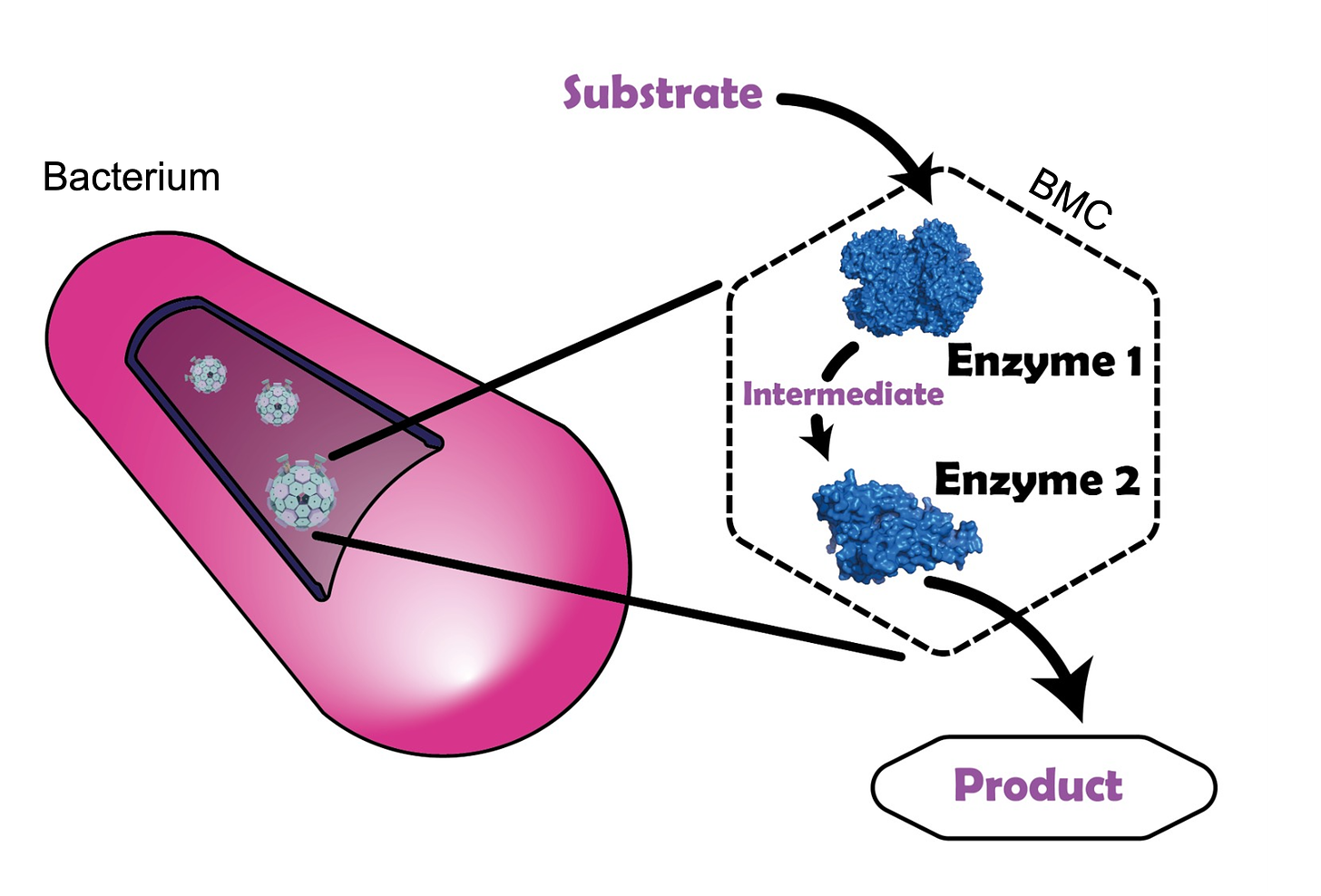
Around the same time, scientists began exploring bacterial microcompartments (BMCs) - protein shells that encapsulate enzymes, acting as rudimentary organelles [10]. BMCs have been successfully reconstituted in various species and repurposed for applications like metabolic engineering and protein encapsulation, showing glimpses of what’s possible [11, 12].
While promising, BMCs are nanoscale structures that don’t quite fit the "organelle" definition we’re using here. BMCs float around in the cytoplasm, processing small molecules but barely exchanging large biomolecules with the external environment. As a result, they behave more like encapsulated complexes with rigid composition, rather than fully programmable, flexible organelle systems. This limited their utility for more complex functions and applications.
Synthetic Organelles: The Future of Cellular Engineering
The promise of synthetic organelles lies in giving bacteria the ability to compartmentalize tasks into neatly defined areas, without interference from other cellular processes.
Creating synthetic organelles in bacteria could revolutionize biotechnology, enabling more complex chemical reactions to occur with greater efficiency. Imagine bacteria that can produce high-value proteins, break down pollutants faster, or manufacture biofuels more sustainably. This isn’t just about copying what eukaryotic cells do - it’s about designing systems that meet the needs of modern synthetic biology.
Moreover, building synthetic organelles would deepen our understanding of how organelles are formed and inherited, opening new avenues for studying the origins of eukaryotic life.
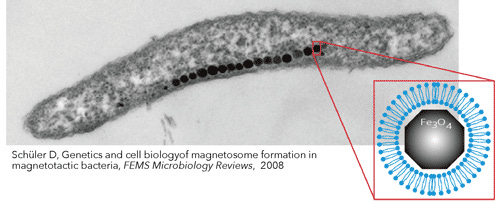
The biggest challenge? Creating organelles that don’t just float around aimlessly, as eukaryotic organelles often do. Biogenesis - the process by which organelles form - remains a mystery for many structures. For example, attempts to transfer magnetosomes from magnetotactic bacteria to E. coli by inserting the gene cluster have been largely unsuccessful [13, 14]. This highlights that even with key genes identified, creating functional organelles is no easy task without fully understanding the underlying mechanisms [15].
However, recent breakthroughs are changing this. New mechanisms for organelle biogenesis are emerging, making it possible to build a new class of synthetic organelles that resemble eukaryotic ones. Next in the series, we’ll explore these exciting strategies, from molecular scaffolds to membraneless organelles powered by liquid-liquid phase separation.
Reference
https://en.wikipedia.org/wiki/Organelle
Greening, Chris, and Trevor Lithgow. "Formation and function of bacterial organelles." Nature Reviews Microbiology 18.12 (2020): 677-689.
Milo, Ron. "What is the total number of protein molecules per cell volume? A call to rethink some published values." Bioessays 35.12 (2013): 1050-1055.
https://en.wikipedia.org/wiki/Fusion_protein
https://en.wikipedia.org/wiki/Green_fluorescent_protein
Castellana, Michele, et al. "Enzyme clustering accelerates processing of intermediates through metabolic channeling." Nature biotechnology 32.10 (2014): 1011-1018.
Dueber, John E., et al. "Synthetic protein scaffolds provide modular control over metabolic flux." Nature biotechnology 27.8 (2009): 753-759.
Delebecque, Camille J., et al. "Organization of intracellular reactions with rationally designed RNA assemblies." Science 333.6041 (2011): 470-474.
Conrado, Robert J., et al. "DNA-guided assembly of biosynthetic pathways promotes improved catalytic efficiency." Nucleic acids research 40.4 (2012): 1879-1889.
https://en.wikipedia.org/wiki/Bacterial_microcompartment
Lau, Yu Heng, et al. "Prokaryotic nanocompartments form synthetic organelles in a eukaryote." Nature communications 9.1 (2018): 1311.
Sigmund, Felix, et al. "Bacterial encapsulins as orthogonal compartments for mammalian cell engineering." Nature communications 9.1 (2018): 1990.
https://en.wikipedia.org/wiki/Magnetosome
Search keywords: "Magnetosome iGEM"; "Magnetosome E. coli", "Magnetosome Synthetic biology"
Uebe, René, and Dirk Schüler. "Magnetosome biogenesis in magnetotactic bacteria." Nature Reviews Microbiology 14.10 (2016): 621-637.
Note: Haotian Guo is the founder and CEO of Ailurus Bio.
Ailurus is a pioneering biocomputer company programming biology as living smart devices, with products like PandaPure® that streamline protein expression and purification directly within cells, eliminating the need for columns or beads. Our mission is to make biology a general-purpose technology - easy to use and as accessible as modern computers. See more at https://www.ailurus.bio





Wonderful piece on synthetic organelles. Looking forward to this series.